The goal of CASP experiment is to assess the state of the art in protein structure prediction via objective blind testing. During over decade, by providing room for community members who are studying what proteins really are, CASP is one of prominent "academic competitions".
Here, I briefly explain the main idea for predicting. The idea was tested at CASP7 that I participated the competition as a member of team ROKKO and ROKKY (Kobe Univ.). Thus, some of descriptions in here involves the research I did at Kobe univ. And also, it includes results that have done by other team members.
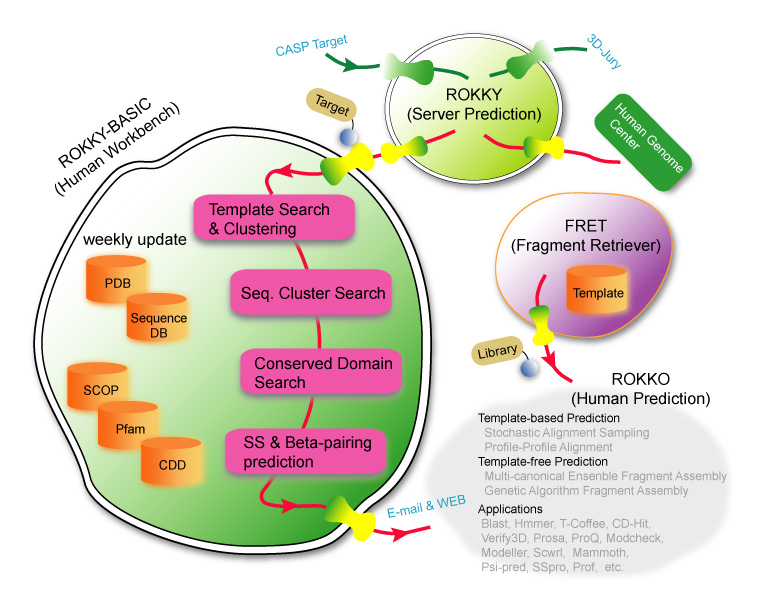
We performed the template-based procedure for predicting targets that have significant PSI-BLAST E-value (< 0.001) or 3D-jury jscore (>50.0). For all remaining targets, we conducted fragment assembly(FA) folding simulation with the different types of fragment libraries. When there exist long alignment gaps (>20 residues) or probably unseen domains (e.g. T0311, T0347, etc.), we first predicted a full-length model with a template and ran FA to predict these broken regions. For a target that is likely to have multiple domains, we parsed it into monomers based on domain DBs, and combined them into a single chain by FA. 13 targets were predicted by the consensus of two FA method (Multi-canonical Ensemble and Genetic Algorithm); by using cluster analysis and visual inspection, we selected five models from independently sampled models by each FA method.
Zscore of CASP7 Target - Template
by CE
by MAMMOTH
Zscore of CASP7 Domains defined by CASP7 assessors
Energy Function: Fujitsuka et al. Proteins 62, 381-398, 2006
Multi-canonical Ensemble: Chikenji et al. J. Chem. Phys. 119, 6895-6903, 2003
Genetic Algorithm: Park S.J. Genome Informatics, 16(2):104-113, 2005
CASP7 proceedings
Official CASP7 Homepage
Forcasp